RVV separated from OPV
Full analysis
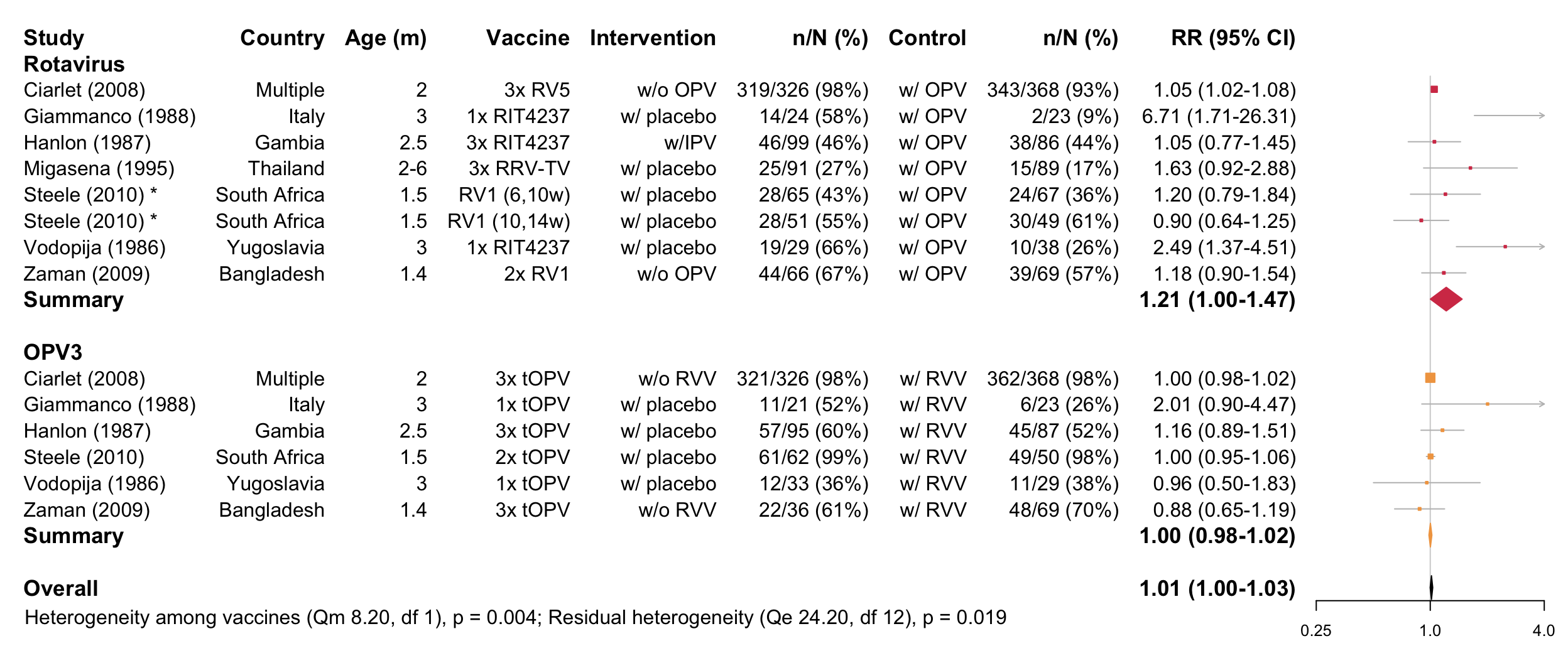
Forest plot
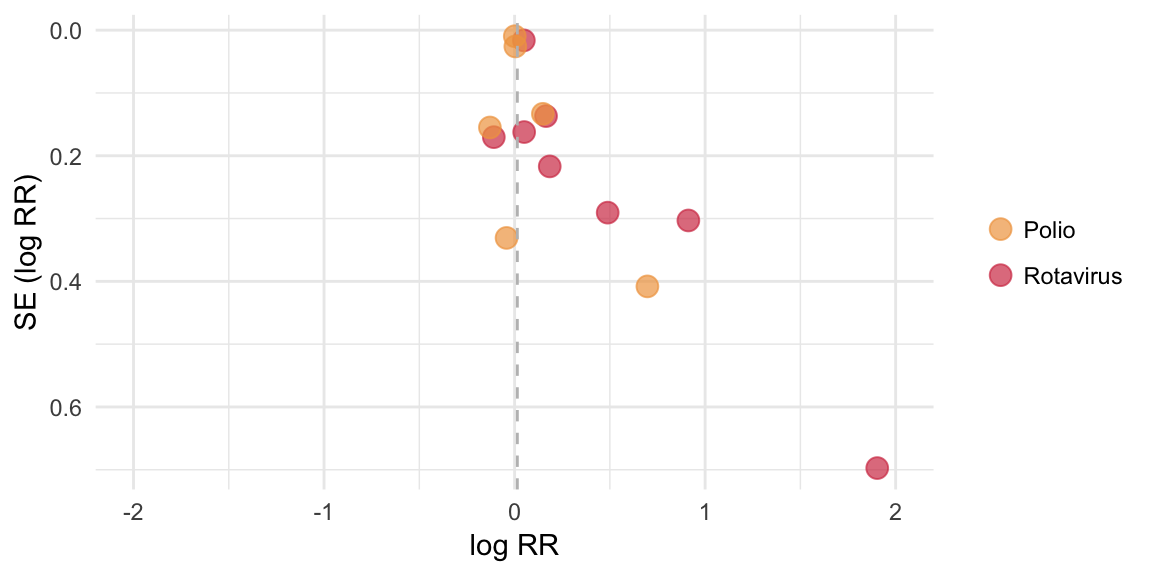
Funnel plot
Egger’s test
## [1] "Infants replicated across vaccines or vaccine effect significant, therefore separate tests performed"
## [1] "Cholera:"
## [1] "Insufficient studies (n<3)"
## [1] "Rotavirus:"
##
## Regression Test for Funnel Plot Asymmetry
##
## model: mixed-effects meta-regression model
## predictor: standard error
##
## test for funnel plot asymmetry: z = 2.8257, p = 0.0047
##
## [1] "OPV3:"
##
## Regression Test for Funnel Plot Asymmetry
##
## model: mixed-effects meta-regression model
## predictor: standard error
##
## test for funnel plot asymmetry: z = 0.9181, p = 0.3586Meta-analysis output
##
## Multivariate Meta-Analysis Model (k = 14; method: REML)
##
## logLik Deviance AIC BIC AICc
## -3.4877 6.9753 10.9753 12.1052 12.1753
##
## Variance Components:
##
## estim sqrt nlvls fixed factor
## sigma^2 0.0000 0.0000 8 no Reference
##
## Test for Heterogeneity:
## Q(df = 13) = 32.4026, p-val = 0.0021
##
## Model Results:
##
## estimate se zval pval ci.lb ci.ub
## 0.0145 0.0078 1.8560 0.0635 -0.0008 0.0299 .
##
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Meta-regression output
##
## Multivariate Meta-Analysis Model (k = 14; method: REML)
##
## Variance Components:
##
## estim sqrt nlvls fixed factor
## sigma^2 0.0000 0.0000 8 no Reference
##
## Test for Residual Heterogeneity:
## QE(df = 12) = 24.1985, p-val = 0.0191
##
## Test of Moderators (coefficient(s) 2):
## QM(df = 1) = 8.2041, p-val = 0.0042
##
## Model Results:
##
## estimate se zval pval ci.lb ci.ub
## intrcpt 0.0019 0.0090 0.2087 0.8347 -0.0157 0.0195
## VaccineRotavirus 0.0524 0.0183 2.8643 0.0042 0.0165 0.0882 **
##
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Intervention-specific outputs
Summary of cholera studies
## [1] "Insufficient studies (n<2)"Summary of rotavirus studies
##
## Random-Effects Model (k = 8; tau^2 estimator: REML)
##
## logLik deviance AIC BIC AICc
## -4.5549 9.1098 13.1098 13.0017 16.1098
##
## tau^2 (estimated amount of total heterogeneity): 0.0382 (SE = 0.0384)
## tau (square root of estimated tau^2 value): 0.1955
## I^2 (total heterogeneity / total variability): 64.64%
## H^2 (total variability / sampling variability): 2.83
##
## Test for Heterogeneity:
## Q(df = 7) = 19.3329, p-val = 0.0072
##
## Model Results:
##
## estimate se zval pval ci.lb ci.ub
## 0.1945 0.0984 1.9778 0.0480 0.0018 0.3873 *
##
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Summary of PV3 studies
##
## Random-Effects Model (k = 6; tau^2 estimator: REML)
##
## logLik deviance AIC BIC AICc
## 3.3313 -6.6626 -2.6626 -3.4438 3.3374
##
## tau^2 (estimated amount of total heterogeneity): 0.0000 (SE = 0.0005)
## tau (square root of estimated tau^2 value): 0.0015
## I^2 (total heterogeneity / total variability): 0.12%
## H^2 (total variability / sampling variability): 1.00
##
## Test for Heterogeneity:
## Q(df = 5) = 4.8656, p-val = 0.4325
##
## Model Results:
##
## estimate se zval pval ci.lb ci.ub
## 0.0019 0.0091 0.2085 0.8348 -0.0159 0.0197
##
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Meta-regression: secondary moderators
Age group
## [1] "No variation in moderator among studies"Income setting
## [1] "Rotavirus-specific analysis performed owing to absence of significant residual heterogeneity for OPV studies"
##
## low_lowermiddle uppermiddle_high
## 2 6##
## Multivariate Meta-Analysis Model (k = 8; method: REML)
##
## Variance Components: none
##
## Test for Residual Heterogeneity:
## QE(df = 6) = 18.9604, p-val = 0.0042
##
## Test of Moderators (coefficient(s) 2):
## QM(df = 1) = 0.3725, p-val = 0.5417
##
## Model Results:
##
## estimate se zval pval
## intrcpt 0.1174 0.1046 1.1220 0.2619
## factor(Income_group)uppermiddle_high -0.0646 0.1058 -0.6103 0.5417
## ci.lb ci.ub
## intrcpt -0.0877 0.3224
## factor(Income_group)uppermiddle_high -0.2720 0.1428
##
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Background immunogenicity (seroconversion rate in the control group)
## [1] "Rotavirus-specific analysis performed owing to absence of significant residual heterogeneity for OPV studies"##
## Multivariate Meta-Analysis Model (k = 8; method: REML)
##
## Variance Components: none
##
## Test for Residual Heterogeneity:
## QE(df = 6) = 13.5770, p-val = 0.0347
##
## Test of Moderators (coefficient(s) 2):
## QM(df = 1) = 5.7559, p-val = 0.0164
##
## Model Results:
##
## estimate se zval pval
## intrcpt 0.4711 0.1745 2.7002 0.0069
## asin(sqrt(Baseline_seroconversion)) -0.3249 0.1354 -2.3991 0.0164
## ci.lb ci.ub
## intrcpt 0.1292 0.8131 **
## asin(sqrt(Baseline_seroconversion)) -0.5904 -0.0595 *
##
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1OPV-specific analysis
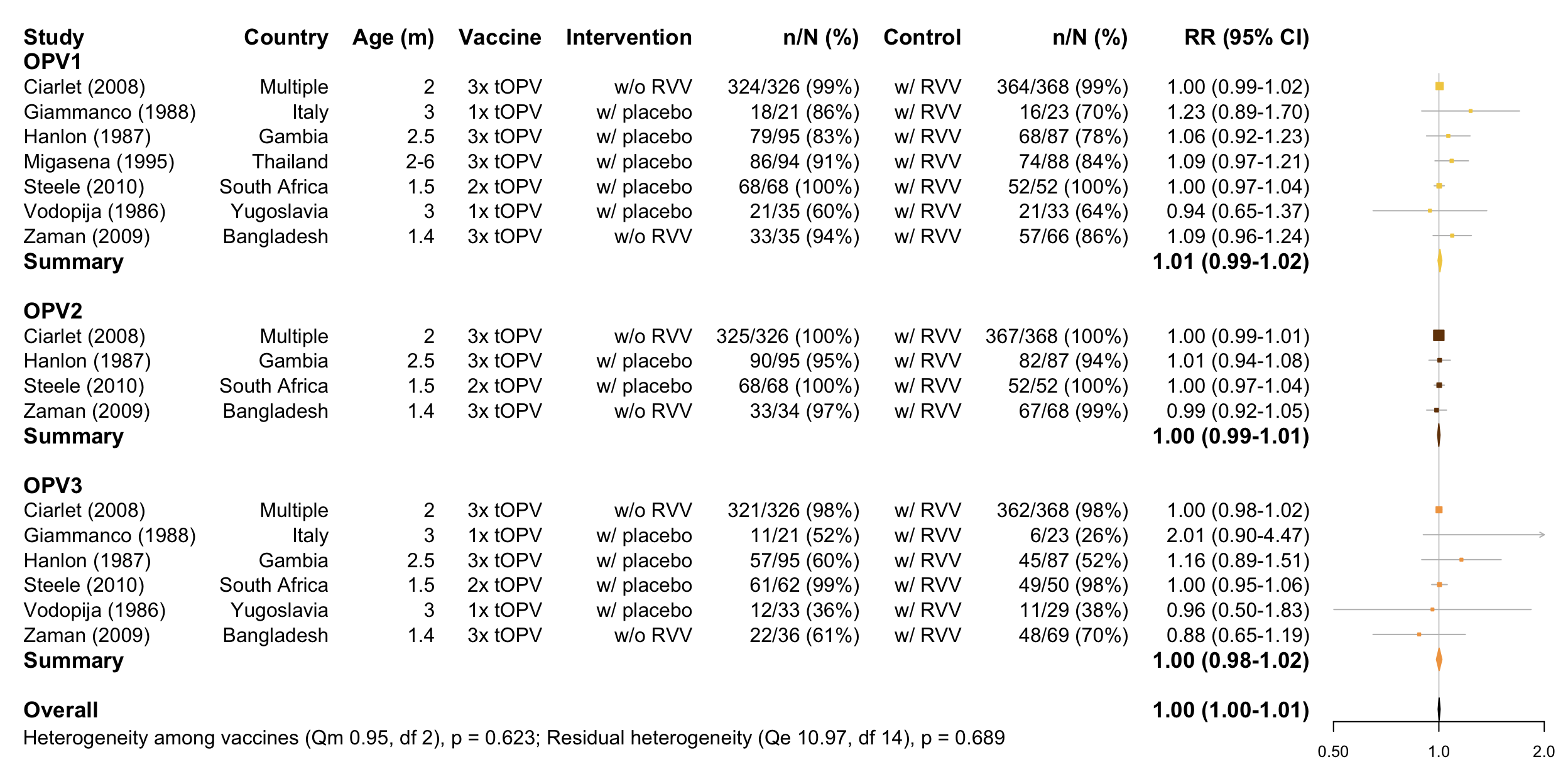
Forest plot
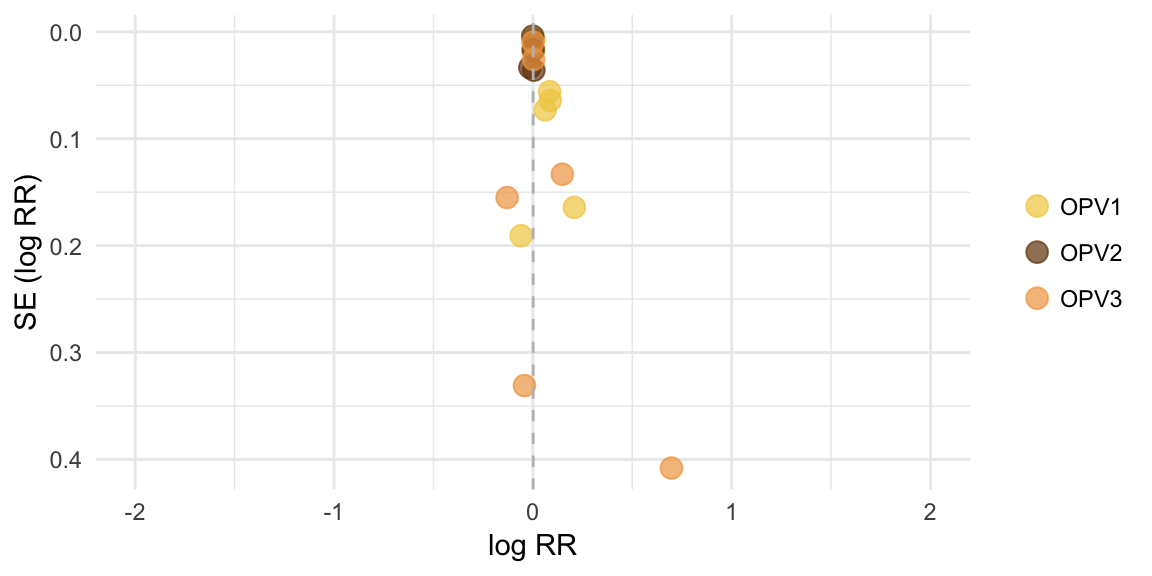
Funnel plot
Egger’s test
Infants replicated across vaccines, therefore separate tests performed
## [1] "OPV1:"##
## Regression Test for Funnel Plot Asymmetry
##
## model: mixed-effects meta-regression model
## predictor: standard error
##
## test for funnel plot asymmetry: z = 1.7901, p = 0.0734## [1] "OPV2:"##
## Regression Test for Funnel Plot Asymmetry
##
## model: mixed-effects meta-regression model
## predictor: standard error
##
## test for funnel plot asymmetry: z = -0.0970, p = 0.9227## [1] "OPV3:"##
## Regression Test for Funnel Plot Asymmetry
##
## model: mixed-effects meta-regression model
## predictor: standard error
##
## test for funnel plot asymmetry: z = 0.9181, p = 0.3586Meta-analysis output
##
## Multivariate Meta-Analysis Model (k = 17; method: REML)
##
## logLik Deviance AIC BIC AICc
## 26.2267 -52.4533 -48.4533 -46.9081 -47.5302
##
## Variance Components:
##
## estim sqrt nlvls fixed factor
## sigma^2 0.0000 0.0000 7 no Reference
##
## Test for Heterogeneity:
## Q(df = 16) = 11.9142, p-val = 0.7499
##
## Model Results:
##
## estimate se zval pval ci.lb ci.ub
## 0.0017 0.0031 0.5392 0.5897 -0.0045 0.0078
##
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Meta-regression output
##
## Multivariate Meta-Analysis Model (k = 17; method: REML)
##
## Variance Components:
##
## estim sqrt nlvls fixed factor
## sigma^2 0.0000 0.0000 7 no Reference
##
## Test for Residual Heterogeneity:
## QE(df = 14) = 10.9686, p-val = 0.6885
##
## Test of Moderators (coefficient(s) 2:3):
## QM(df = 2) = 0.9456, p-val = 0.6232
##
## Model Results:
##
## estimate se zval pval ci.lb ci.ub
## intrcpt 0.0069 0.0063 1.0885 0.2764 -0.0055 0.0194
## Measure_of_SCOPV2 N-AB -0.0073 0.0075 -0.9722 0.3310 -0.0219 0.0074
## Measure_of_SCOPV3 N-AB -0.0050 0.0110 -0.4574 0.6474 -0.0266 0.0165
##
## intrcpt
## Measure_of_SCOPV2 N-AB
## Measure_of_SCOPV3 N-AB
##
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Serotype-specific outputs
Summary of PV1 studies
##
## Random-Effects Model (k = 7; tau^2 estimator: REML)
##
## logLik deviance AIC BIC AICc
## 8.2203 -16.4405 -12.4405 -12.8570 -8.4405
##
## tau^2 (estimated amount of total heterogeneity): 0 (SE = 0.0002)
## tau (square root of estimated tau^2 value): 0
## I^2 (total heterogeneity / total variability): 0.00%
## H^2 (total variability / sampling variability): 1.00
##
## Test for Heterogeneity:
## Q(df = 6) = 5.8625, p-val = 0.4388
##
## Model Results:
##
## estimate se zval pval ci.lb ci.ub
## 0.0069 0.0063 1.0885 0.2764 -0.0055 0.0194
##
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Summary of PV2 studies
##
## Random-Effects Model (k = 4; tau^2 estimator: REML)
##
## logLik deviance AIC BIC AICc
## 8.5846 -17.1691 -13.1691 -14.9719 -1.1691
##
## tau^2 (estimated amount of total heterogeneity): 0 (SE = 0.0002)
## tau (square root of estimated tau^2 value): 0
## I^2 (total heterogeneity / total variability): 0.00%
## H^2 (total variability / sampling variability): 1.00
##
## Test for Heterogeneity:
## Q(df = 3) = 0.2404, p-val = 0.9708
##
## Model Results:
##
## estimate se zval pval ci.lb ci.ub
## -0.0004 0.0039 -0.0892 0.9289 -0.0081 0.0074
##
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Summary of PV3 studies
##
## Random-Effects Model (k = 6; tau^2 estimator: REML)
##
## logLik deviance AIC BIC AICc
## 3.3313 -6.6626 -2.6626 -3.4438 3.3374
##
## tau^2 (estimated amount of total heterogeneity): 0.0000 (SE = 0.0005)
## tau (square root of estimated tau^2 value): 0.0015
## I^2 (total heterogeneity / total variability): 0.12%
## H^2 (total variability / sampling variability): 1.00
##
## Test for Heterogeneity:
## Q(df = 5) = 4.8656, p-val = 0.4325
##
## Model Results:
##
## estimate se zval pval ci.lb ci.ub
## 0.0019 0.0091 0.2085 0.8348 -0.0159 0.0197
##
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1